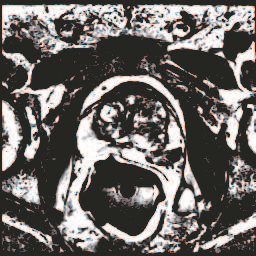
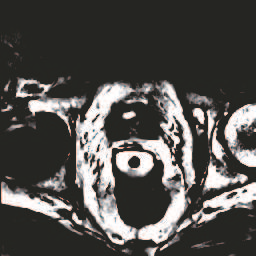
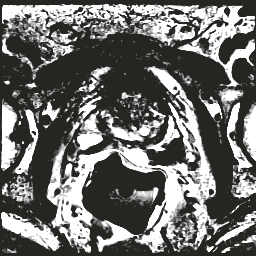
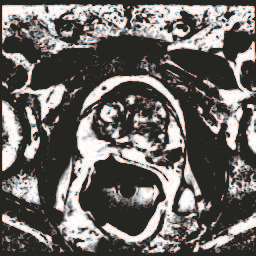
In this paper, we focus on three problems in deep learning based medical image segmentation. Firstly, U-net, as a popular model for medical image segmentation, is difficult to train when convolutional layers increase even though a deeper network usually has a better generalization ability because of more learnable parameters. Secondly, the exponential ReLU (ELU), as an alternative of ReLU, is not much different from ReLU when the network of interest gets deep. Thirdly, the Dice loss, as one of the pervasive loss functions for medical image segmentation, is not effective when the prediction is close to ground truth and will cause oscillation during training. To address the aforementioned three problems, we propose and validate a deeper network that can fit medical image datasets that are usually small in the sample size. Meanwhile, we propose a new loss function to accelerate the learning process and a combination of different activation functions to improve the network performance. Our experimental results suggest that our network is comparable or superior to state-of-the-art methods.
翻译:在本文中,我们集中关注基于深入学习的医学图像分割的三个问题。 首先,U-net作为医学图像分割的流行模型,很难在进化层增加时进行培训,尽管更深的网络由于学习的参数而通常具有更好的概括性能力。 其次,指数性ReLU(ELU)作为RELU的替代品,在网络深入时与RELU没有多大区别。 第三,当医学图像分割的普遍损失功能之一,当预测接近地面真相并会在培训期间引起振荡时,Dice损失是无效的。为了解决上述三个问题,我们提议并验证一个更深的网络,这个网络能够适应医学图像数据集,该数据集通常在抽样规模上很小。与此同时,我们提议一个新的损失功能来加速学习过程,并结合不同的激活功能来改善网络的性能。我们的实验结果表明,我们的网络与最先进的方法相比,或者更优越。