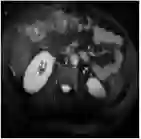
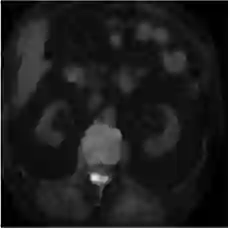
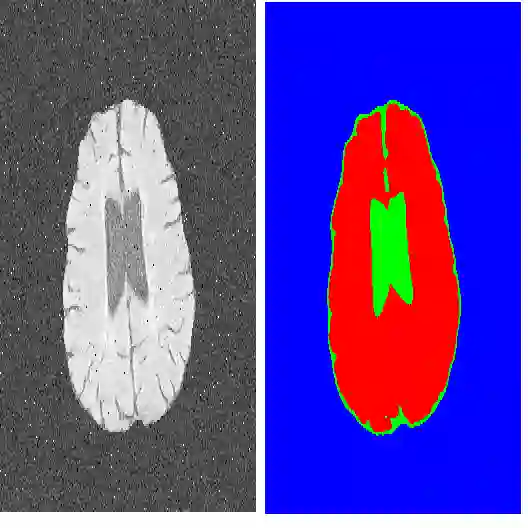
This work presents a novel framework CISFA (Contrastive Image synthesis and Self-supervised Feature Adaptation)that builds on image domain translation and unsupervised feature adaptation for cross-modality biomedical image segmentation. Different from existing works, we use a one-sided generative model and add a weighted patch-wise contrastive loss between sampled patches of the input image and the corresponding synthetic image, which serves as shape constraints. Moreover, we notice that the generated images and input images share similar structural information but are in different modalities. As such, we enforce contrastive losses on the generated images and the input images to train the encoder of a segmentation model to minimize the discrepancy between paired images in the learned embedding space. Compared with existing works that rely on adversarial learning for feature adaptation, such a method enables the encoder to learn domain-independent features in a more explicit way. We extensively evaluate our methods on segmentation tasks containing CT and MRI images for abdominal cavities and whole hearts. Experimental results show that the proposed framework not only outputs synthetic images with less distortion of organ shapes, but also outperforms state-of-the-art domain adaptation methods by a large margin.
翻译:这项工作提出了一个新的框架 CISFA( CISFA (Contratsive 图像合成和自监督的功能适应), 该框架以图像域翻译和未经监督的特性适应为基础, 用于跨现代生物医学图像分割。 不同于现有的作品, 我们使用单向基因模型, 并添加了输入图像抽样片段和相应的合成图像之间的加权、 偏斜的对比性损失, 用作形状限制。 此外, 我们注意到, 生成的图像和输入图像共享类似的结构信息, 但形式不同 。 因此, 我们强制对生成的图像和输入图像进行对比性损失, 以训练分解模型的编码器, 以尽量减少所学嵌入空间中配对图像之间的差异 。 与目前依靠对抗性学习进行特征适应的工程相比, 这样的方法使编码器能够以更清晰的方式学习与域独立的特征特征。 我们广泛评估了我们关于含有C 和 MRI 图像的分解任务的方法, 用于腹腔和整个心脏 。 实验结果显示, 拟议的框架不仅输出器官形状的合成图象,, 而且还以大比例法 外形 。