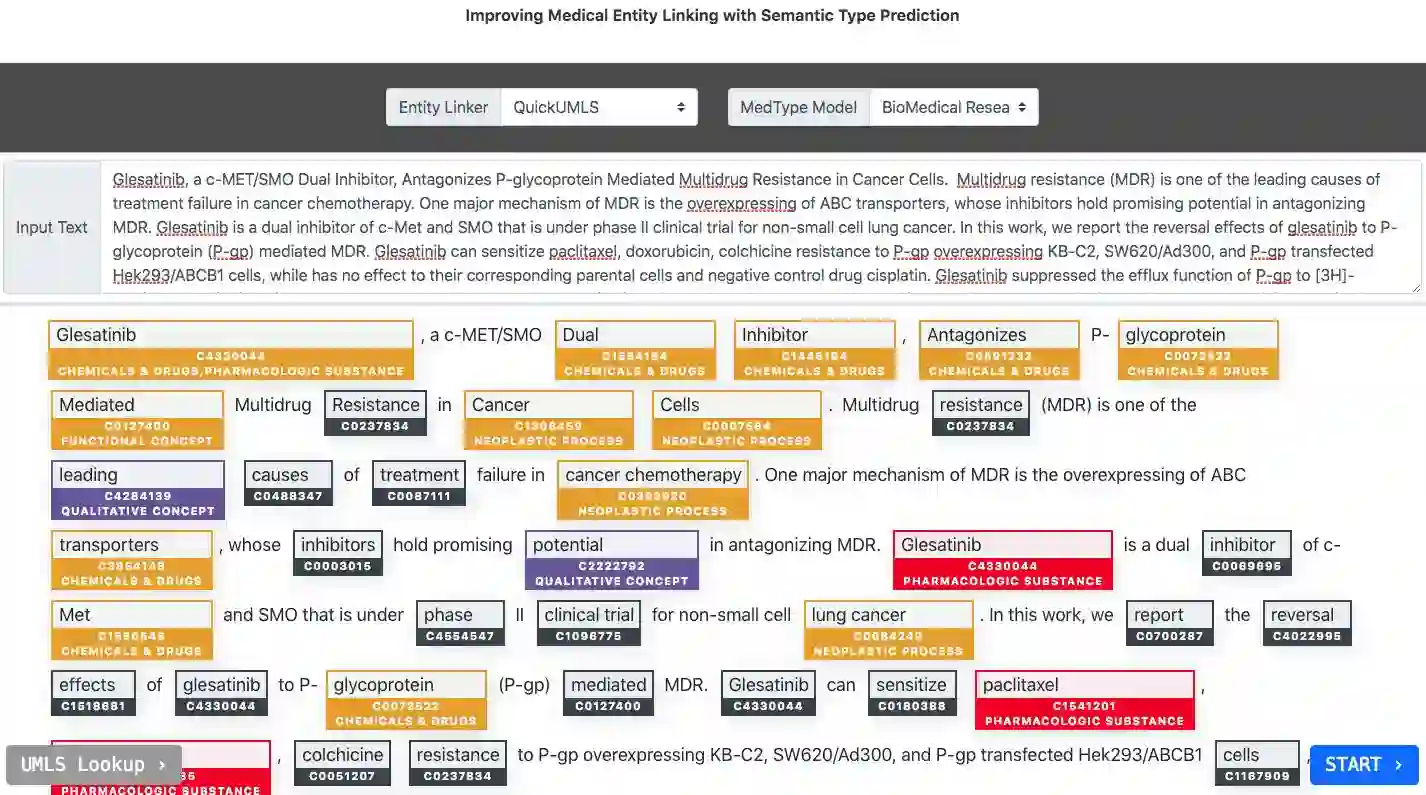
Pre-trained language models induce dense entity representations that offer strong performance on entity-centric NLP tasks, but such representations are not immediately interpretable. This can be a barrier to model uptake in important domains such as biomedicine. There has been recent work on general interpretable representation learning (Onoe and Durrett, 2020), but these domain-agnostic representations do not readily transfer to the important domain of biomedicine. In this paper, we create a new entity type system and training set from a large corpus of biomedical texts by mapping entities to concepts in a medical ontology, and from these to Wikipedia pages whose categories are our types. From this mapping we derive Biomedical Interpretable Entity Representations(BIERs), in which dimensions correspond to fine-grained entity types, and values are predicted probabilities that a given entity is of the corresponding type. We propose a novel method that exploits BIER's final sparse and intermediate dense representations to facilitate model and entity type debugging. We show that BIERs achieve strong performance in biomedical tasks including named entity disambiguation and entity label classification, and we provide error analysis to highlight the utility of their interpretability, particularly in low-supervision settings. Finally, we provide our induced 68K biomedical type system, the corresponding 37 million triples of derived data used to train BIER models and our best performing model.
翻译:培训前语言模式导致实体代表密集,在以实体为中心的NLP任务上表现良好,但这种代表不能立即解释。这可能成为生物医学等重要领域模型吸收的障碍。最近开展了关于通用解释代表性学习的工作(Onoe和Durrett,2020年),但这些域名代表并不轻易转移到生物医学的重要领域。在本文件中,我们建立了一个新的实体类型系统和培训,从大量生物医学文本中,通过绘制实体图谱,到医学本体学的概念,从这些实体到属于我们类别的概念,到维基百科网页,这可能成为生物医学中间实体代表(BIERs)在生物医学重要领域采用模型方面的障碍。我们从这一绘图中得出生物医学中间代表(BIERs)与微分化实体类型相对应,而数值是预测某一实体属于相应类型的概率的概率。我们提出了一个新的方法,即利用BIER公司最后的稀薄和中间密集的表达方式,以便利模型和实体型号调试。我们表明,BIERs在生物医学任务中取得了强有力的表现,特别是实体分辨和实体标签分类,我们提供的推算出38的模型的模型,我们最后提供了用于BER-38型模型的模型的模型的模型的模型的模型。我们提供的模型的导导导导导数据。