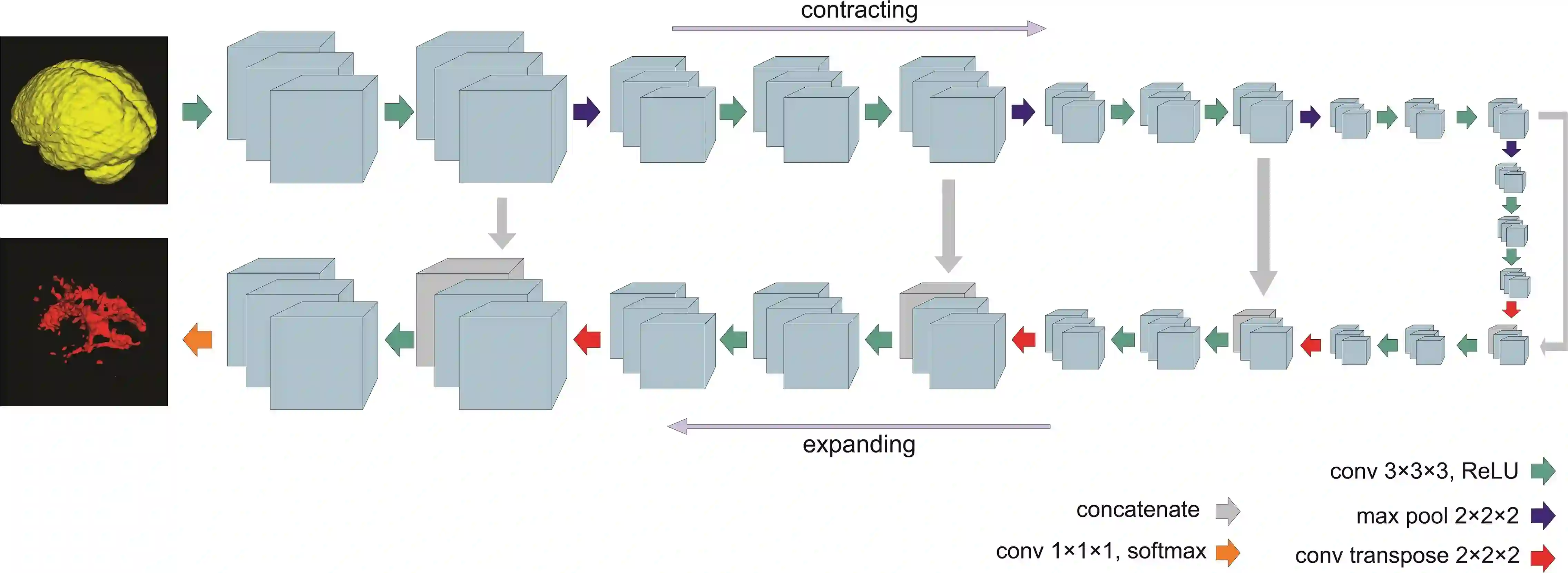
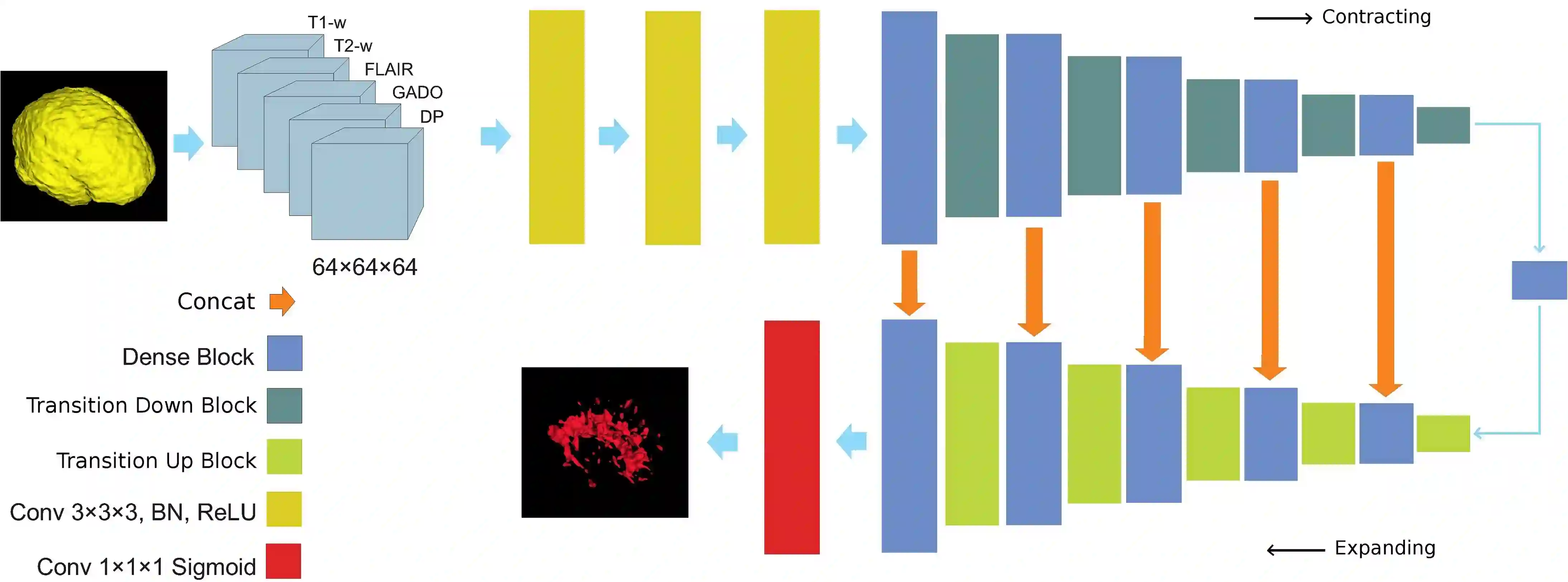
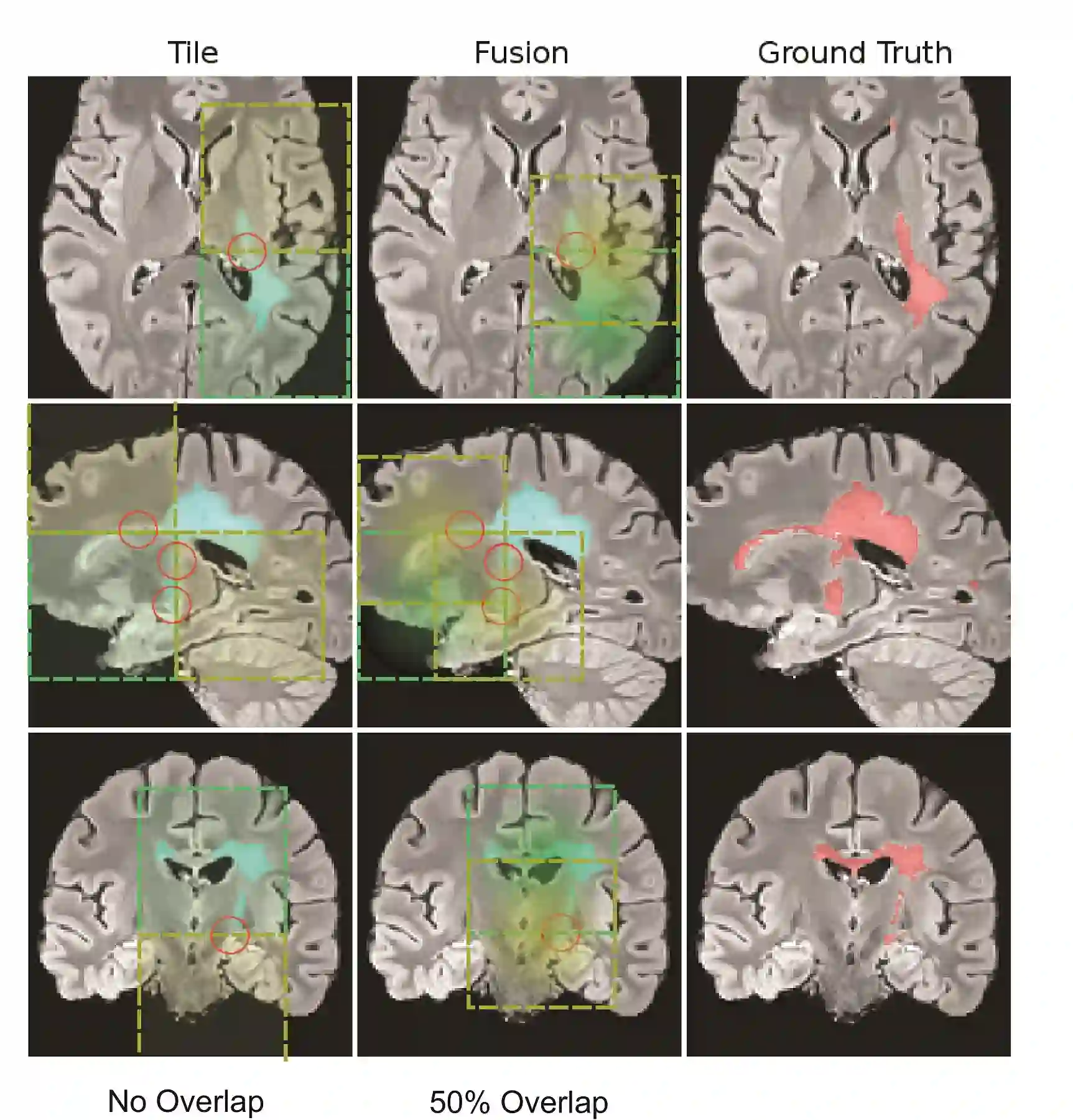
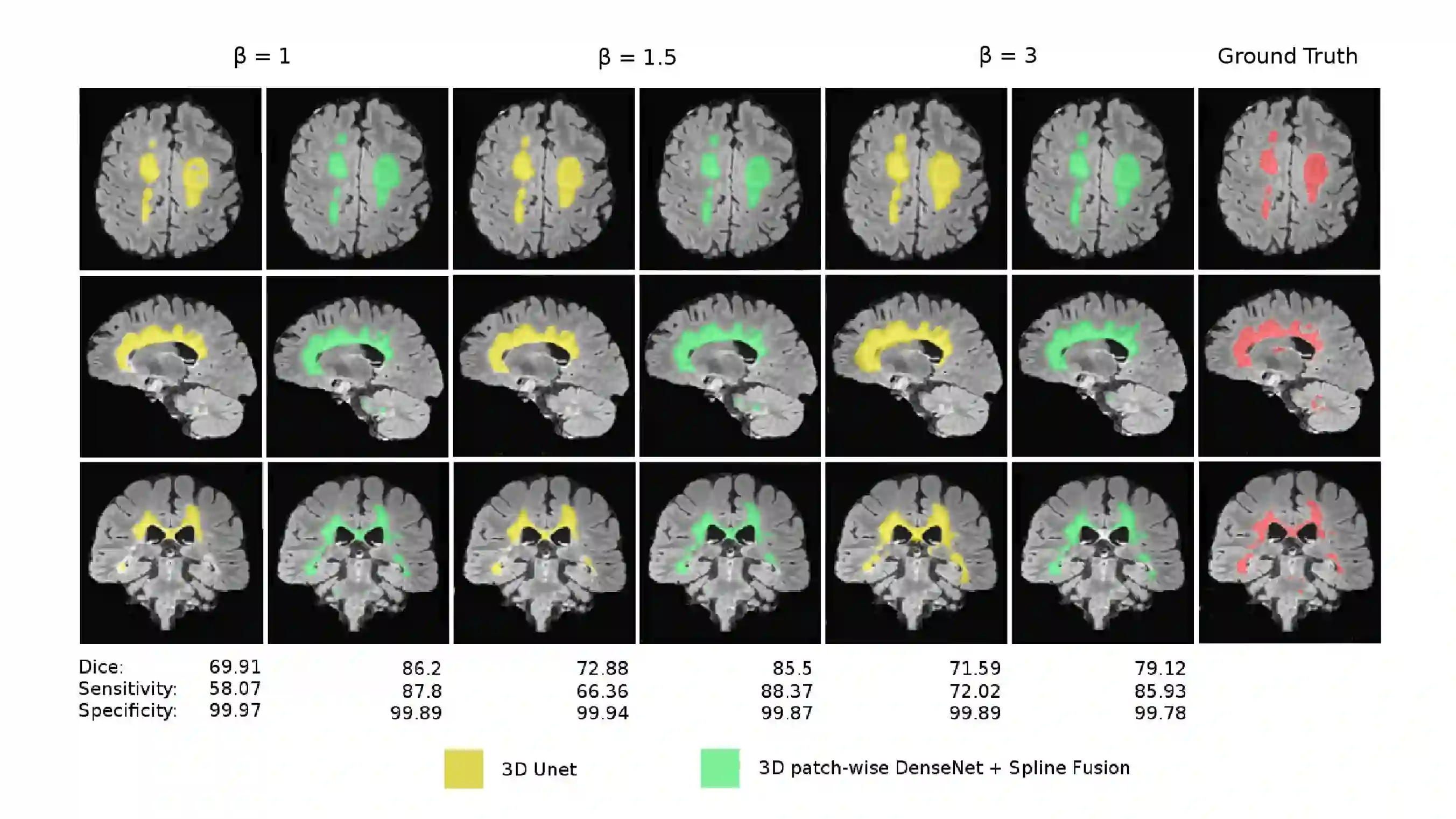
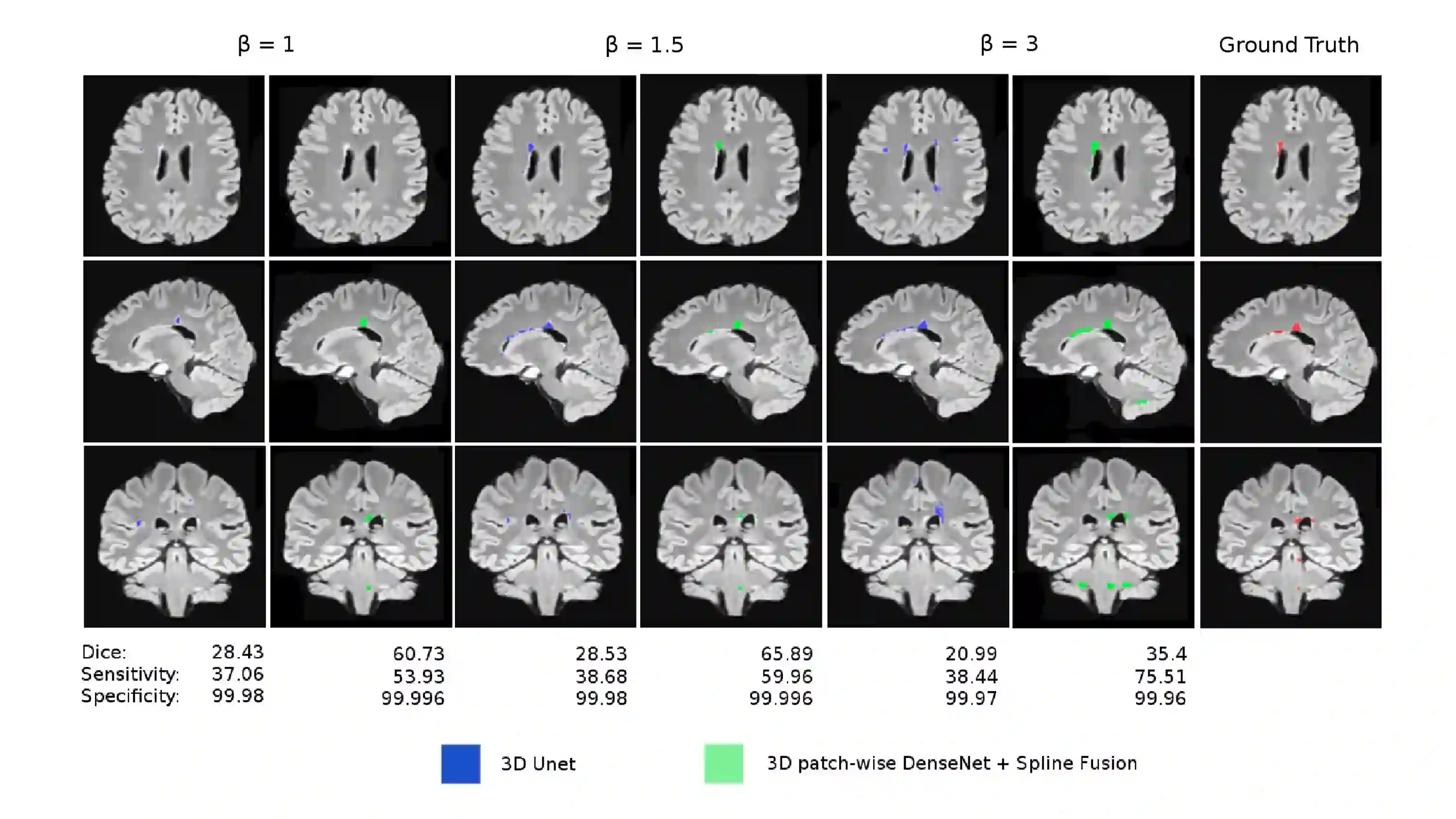
Fully convolutional deep neural networks have been asserted to be fast and precise frameworks with great potential in image segmentation. One of the major challenges in utilizing such networks raises when data is unbalanced, which is common in many medical imaging applications such as lesion segmentation where lesion class voxels are often much lower in numbers than non-lesion voxels. A trained network with unbalanced data may make predictions with high precision and low recall, being severely biased towards the non-lesion class which is particularly undesired in medical applications where false negatives are actually more important than false positives. Various methods have been proposed to address this problem including two step training, sample re-weighting, balanced sampling, and similarity loss functions. In this paper we developed a patch-wise 3D densely connected network with an asymmetric loss function, where we used large overlapping image patches for intrinsic and extrinsic data augmentation, a patch selection algorithm, and a patch prediction fusion strategy based on B-spline weighted soft voting to take into account the uncertainty of prediction in patch borders. We applied this method to lesion segmentation based on the MSSEG 2016 and ISBI 2015 challenges, where we achieved average Dice similarity coefficient of 69.9% and 65.74%, respectively. In addition to the proposed loss, we trained our network with focal and generalized Dice loss functions. Significant improvement in $F_1$ and $F_2$ scores and the APR curve was achieved in test using the asymmetric similarity loss layer and our 3D patch prediction fusion. The asymmetric similarity loss based on $F_\beta$ scores generalizes the Dice similarity coefficient and can be effectively used with the patch-wise strategy developed here to train fully convolutional deep neural networks for highly unbalanced image segmentation.
翻译:完全深层神经网络被证明是快速和精确的框架,在图像分割方面潜力巨大。当数据不平衡时,使用这种网络的主要挑战之一就会产生。数据不平衡时,这是许多医学成像应用中常见的一个挑战,例如,腐蚀类氧化物在数量上往往比非腐蚀素低得多。一个经过培训的不平衡数据网络可能会做出高度精确和低回调的预测,严重偏向于非遗赠类,在医疗应用程序中,虚假的正值比假的正值更重要,在使用这种网络时,使用各种方法解决这一问题,包括两步制培训、抽样重整、平衡的取样和类似的损失功能。在本文中,我们开发了一个偏差的三维紧密连通网络,其中我们使用大量重叠的图像补差来增强内脏和外脏数据,一个补差选择算法,以及基于B-spline加权的软票计算,以考虑补差边界预测的不确定性。我们应用了这种方法来解决这个问题,包括两步制的精度、抽样再配比重的货币网络的三维值重的三维度,我们用了MSS-E-eal-eal-IL 和ILLL IM Creal 和IL