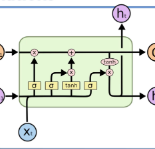
Researchers working on computational analysis of Whole Slide Images (WSIs) in histopathology have primarily resorted to patch-based modelling due to large resolution of each WSI. The large resolution makes WSIs infeasible to be fed directly into the machine learning models due to computational constraints. However, due to patch-based analysis, most of the current methods fail to exploit the underlying spatial relationship among the patches. In our work, we have tried to integrate this relationship along with feature-based correlation among the extracted patches from the particular tumorous region. For the given task of classification, we have used BiLSTMs to model both forward and backward contextual relationship. RNN based models eliminate the limitation of sequence size by allowing the modelling of variable size images within a deep learning model. We have also incorporated the effect of spatial continuity by exploring different scanning techniques used to sample patches. To establish the efficiency of our approach, we trained and tested our model on two datasets, microscopy images and WSI tumour regions. After comparing with contemporary literature we achieved the better performance with accuracy of 90% for microscopy image dataset. For WSI tumour region dataset, we compared the classification results with deep learning networks such as ResNet, DenseNet, and InceptionV3 using maximum voting technique. We achieved the highest performance accuracy of 84%. We found out that BiLSTMs with CNN features have performed much better in modelling patches into an end-to-end Image classification network. Additionally, the variable dimensions of WSI tumour regions were used for classification without the need for resizing. This suggests that our method is independent of tumour image size and can process large dimensional images without losing the resolution details.
翻译:在组织病理学中,从事整体幻灯片图像计算分析的研究者主要采用基于补丁的建模方法,因为每部图像的分辨率很大。大型分辨率使得由于计算限制,无法直接输入机器学习模型。然而,由于基于补丁的分析,大多数当前方法未能利用补丁之间的基本空间关系。在我们的工作中,我们试图将这种关系与从特定肿瘤区域提取的补丁间基于地貌的相互关系结合起来。为了完成既定的分类任务,我们使用了BILSTM来模拟前向和后向背景关系。基于RNNN的模型通过允许在深层次学习模型中建模变量大小图像而消除序列大小的限制。由于基于补丁分析,我们还纳入了空间连续性的效果,探索了用于抽样补丁之间的不同扫描技术。为了确定我们的方法效率,我们用两个数据集、显微镜图像和WSI肿瘤区域之间的分类方法。在与现代文献的分类中,我们实现了90%的精确性能,用于显微镜图像的大型图像设置。在使用甚深层次的图像网络中,我们用这种精确度来比较,我们用这种精确度数据网络来学习这种精确度数据。