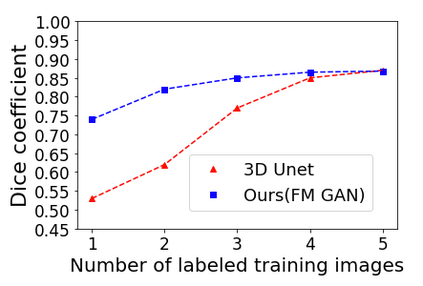
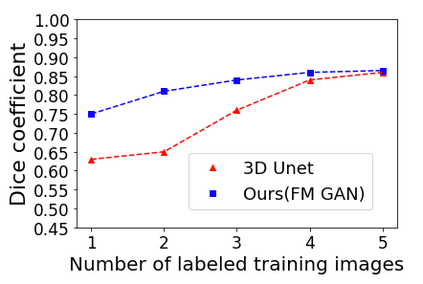
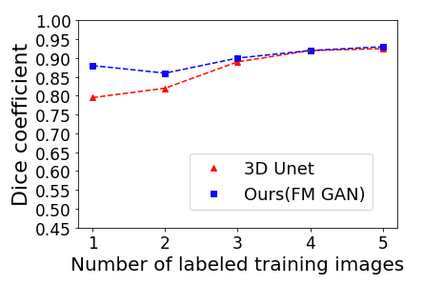
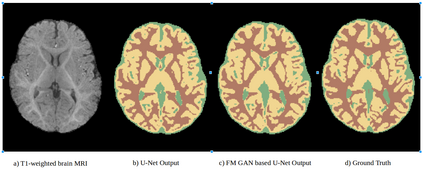
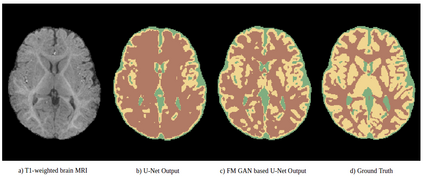
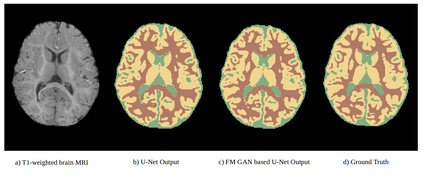
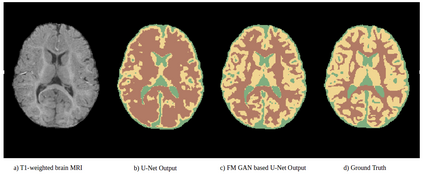
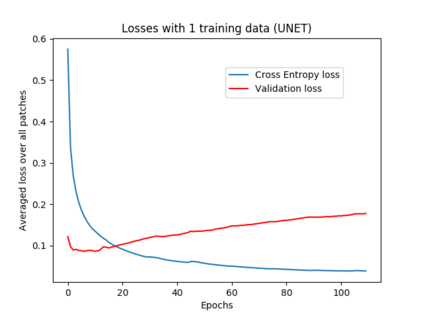
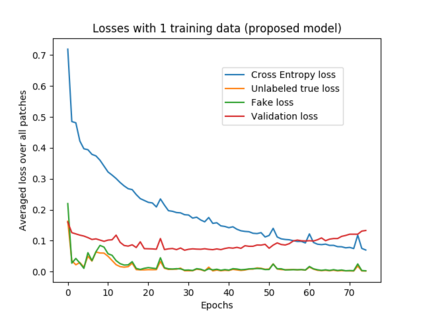
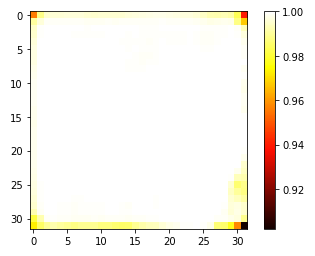
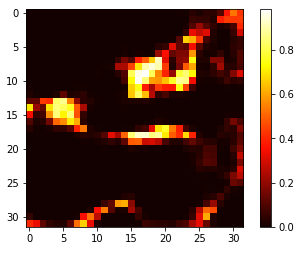
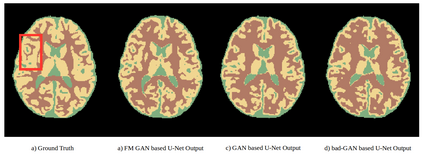
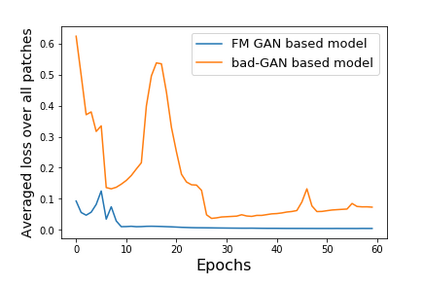
We address the problem of segmenting 3D multi-modal medical images in scenarios where very few labeled examples are available for training. Leveraging the recent success of adversarial learning for semi-supervised segmentation, we propose a novel method based on Generative Adversarial Networks (GANs) to train a segmentation model with both labeled and unlabeled images. The proposed method prevents over-fitting by learning to discriminate between true and fake patches obtained by a generator network. Our work extends current adversarial learning approaches, which focus on 2D single-modality images, to the more challenging context of 3D volumes of multiple modalities. The proposed method is evaluated on the problem of segmenting brain MRI from the iSEG-2017 and MRBrainS 2013 datasets. Significant performance improvement is reported, compared to state-of-art segmentation networks trained in a fully-supervised manner. In addition, our work presents a comprehensive analysis of different GAN architectures for semi-supervised segmentation, showing recent techniques like feature matching to yield a higher performance than conventional adversarial training approaches. Our code is publicly available at https://github.com/arnab39/FewShot_GAN-Unet3D
翻译:我们利用最近对半监督的截肢进行对抗性学习的成功经验,提出了一种基于创用反对流网络(GANs)的新颖方法,以培训带有标签和无标签图像的分解模式;拟议方法防止因学习区分发电机网络获取的真实和假补丁而过度适应。我们的工作将当前以2D单式模式图像为重点的对抗性学习方法扩大到3D多模式中更具挑战性的范围。我们建议的方法是针对iSEG-2017和MRBRAINS 2013数据集对脑MRI进行分解的问题进行评估。报告与以完全监督方式培训的状态分解网络相比,绩效有显著改进。此外,我们的工作还全面分析了半超导分解的不同的GAN结构,展示了与比常规的对立培训方法更高性能相匹配的最近技术。我们的代码在 https://gis/Angu-Annal-D3 上公开提供。