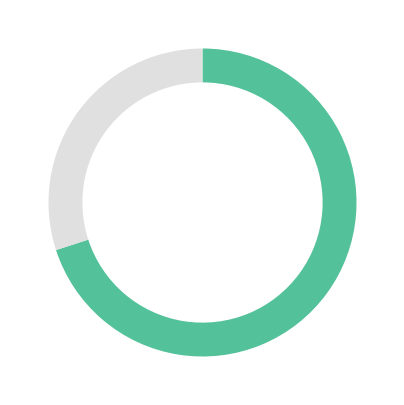
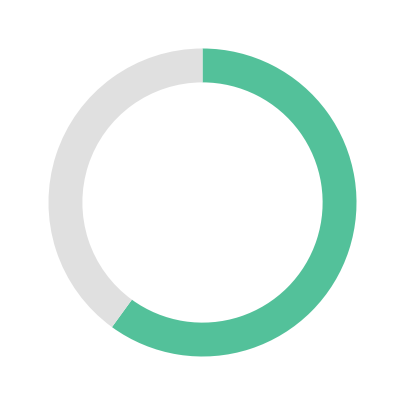
Large Language Models (LLMs) have demonstrated remarkable capabilities on general text; however, their proficiency in specialized scientific domains that require deep, interconnected knowledge remains largely uncharacterized. Metabolomics presents unique challenges with its complex biochemical pathways, heterogeneous identifier systems, and fragmented databases. To systematically evaluate LLM capabilities in this domain, we introduce MetaBench, the first benchmark for metabolomics assessment. Curated from authoritative public resources, MetaBench evaluates five capabilities essential for metabolomics research: knowledge, understanding, grounding, reasoning, and research. Our evaluation of 25 open- and closed-source LLMs reveals distinct performance patterns across metabolomics tasks: while models perform well on text generation tasks, cross-database identifier grounding remains challenging even with retrieval augmentation. Model performance also decreases on long-tail metabolites with sparse annotations. With MetaBench, we provide essential infrastructure for developing and evaluating metabolomics AI systems, enabling systematic progress toward reliable computational tools for metabolomics research.
翻译:暂无翻译