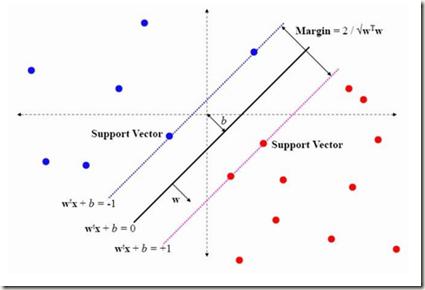
Kernel-based support vector machines (SVMs) are supervised machine learning algorithms for classification and regression problems. We introduce a method to train SVMs on a D-Wave 2000Q quantum annealer and study its performance in comparison to SVMs trained on conventional computers. The method is applied to both synthetic data and real data obtained from biology experiments. We find that the quantum annealer produces an ensemble of different solutions that often generalizes better to unseen data than the single global minimum of an SVM trained on a conventional computer, especially in cases where only limited training data is available. For cases with more training data than currently fits on the quantum annealer, we show that a combination of classifiers for subsets of the data almost always produces stronger joint classifiers than the conventional SVM for the same parameters.
翻译:基于内核的支持矢量机器(SVMs)是监督分类和回归问题的机器学习算法(SVMs),我们引入了一种方法,用D-Wave 2000Q量子annealer来培训SVMs,并与常规计算机培训的SVMs相比,研究其性能;这种方法既适用于合成数据,也适用于从生物学实验中获得的真实数据;我们发现量子annealer产生的不同解决办法组合,往往比用常规计算机培训的SVM的单一全球最低值更好地概括为不可见数据,特别是在只有有限的培训数据的情况下;对于培训数据比目前量子anealer更适合的培训数据的情况,我们表明,对于同一参数而言,数据子组的分类器组合几乎总是产生比常规SVM更强大的联合分类器。